Placement Algorithms
Placement Algorithms
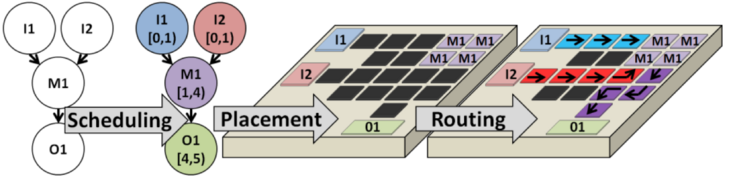
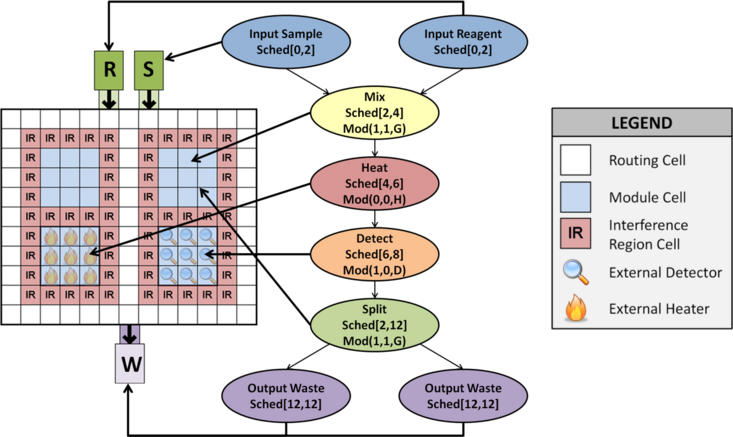
This page summarizes the DMFB Placement algorithms that have been implemented and integrated into the framework, and cites the papers that describe them in detail. Placement determines the precise locations on the DMFB surface at which assay operations are performed at each time-step. The placer must ensure appropriate spacing between operations to prevent cross-contamination, and must limit fragmentation to the fullest extent possible.
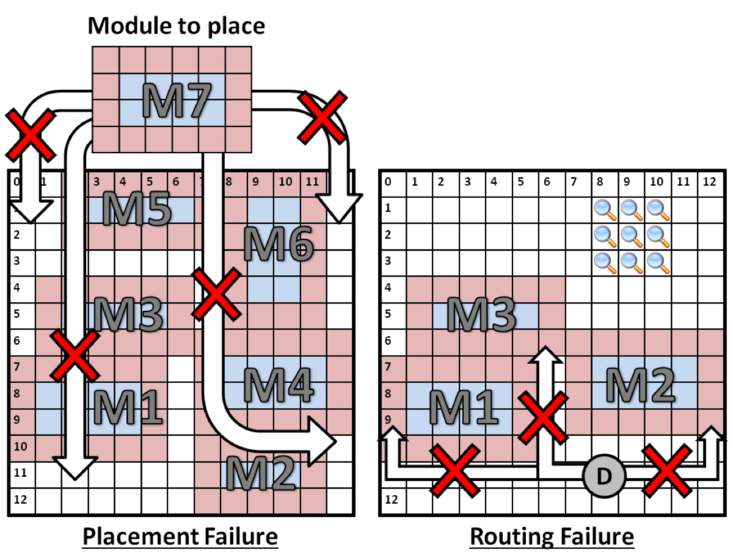
Placement is a NP-complete problem. As shown in the following example, placement may fail due to fragmentation (left): even though there is sufficient area on-chip to fit module M7, it is not available as a contiguous 4x6 free region; likewise, placement may cause routing failures (right): here, several abutted modules block all paths from droplet D's current position to the detector on the opposite side of the chip.
Keep All Maximal Empty Rectangles (KAMER)
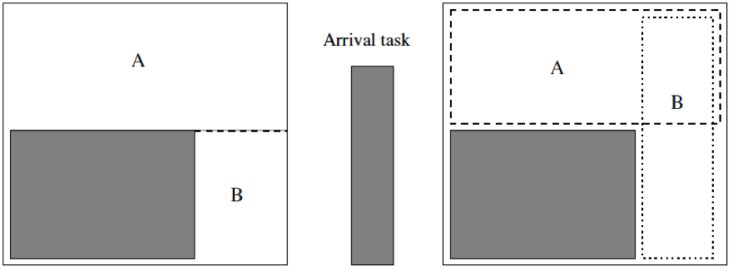
KAMER is a greedy 2D placement algorithm that tries to maximize the amount of contiguous free space that remains after each module is placed. Free space s represented in terms of overlapping rectangles of maximal size, as shown below. KAMER was originally proposed as a solution for dynamic task placement for runtime reconfigurable spatial computing devices; it turns out that the problem formulation perfect fits the needs of DMFBs as well.
We used KAMER as a baseline for comparison in the following two papers:
D. Grissom and P. Brisk
Fast Online Synthesis of Digital Microfluidic Biochips
IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems (TCAD)
33(3):356-369, February, 2014
Paper
D. Grissom and P. Brisk
Fast Online Synthesis of Generally Programmable Digital Microfluidic Biochips
10th IEEE/ACM/IFIP International Conference on Hardware/Software Codesign and System Synthesis (CODES-ISSS)
Tampere, Finland, October 7-12, 2012, pp. 413-422
Paper Slides
KAMER was originally introduced in the following paper:
K. Bazargan, R. Kastner, and M. Sarrafzadeh
Fast Template Placement for Reconfigurable Computing Systems
IEEE Design & Test of Computers
17(1):68-83, January-March, 2000
Our implementation of KAMER uses a data structure described in the following paper to represent free space:
Y. Lu, T. Marconi, G. Gaydadjiev, and K. Bertels
An Efficient Algorithm for Free Resources Management on the FPGA
Design Automation and Test in Europe (DATE)
Munich, Germany, March 10-14, 2008, pp. 1095-1098
Virtual Topology Placer
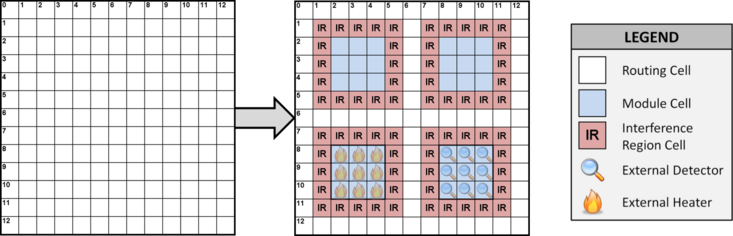
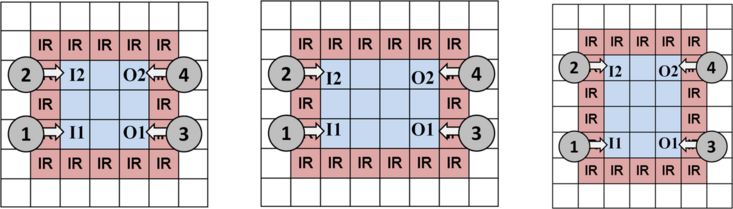
A Virtual Topology determines a-priori the location where assay operations may occur on a DMFB. It logically partitions the DMFB into a set of Work Modules, which perform assay operations {mix, merge, store, split}, and a 2D mesh network of Streets. A subset of work modules is associated with external devices (heaters, detectors, etc.), giving them extra operations. An example is shown below.
The benefits of a virtual topology compared to a traditional placer are as follows:
♦ It clearly articulates the available resources (Work Modules) to the scheduler.
♦ It converts placement into a binding problem, as shown below, which eliminates fragmentation.
♦ It guarantees deadlock-free droplet routing.
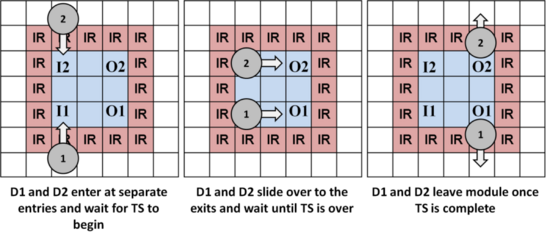
Work Modules can vary in size and shape. One of the keys to ensuring droplet routing is to provide each droplet a dedicated input and output position on the perimeter of the module, as shown below. Although this limits the number of droplets that each module can store, it eliminates the possibility of congestion and deadlock when droplets try to enter and leave a module at the same time. This property (plus the existence of an obstacle-free path between every pair of modules), guarantees the existence of a deadlock-free route.
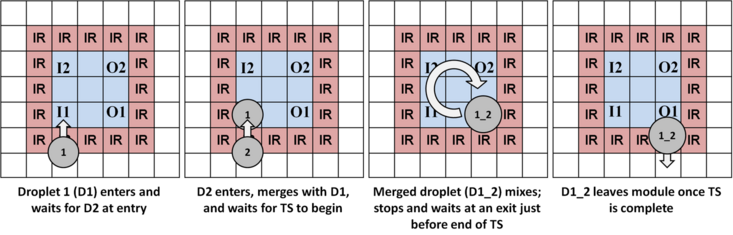
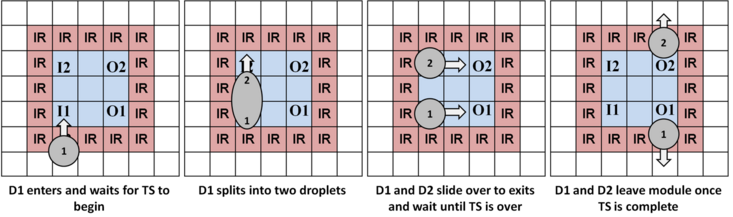
The images below illustrate mixing, splitting, and storage using work modules.
We introduced the virtual topology in the following two papers:
D. Grissom and P. Brisk
Fast Online Synthesis of Digital Microfluidic Biochips
IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems (TCAD)
33(3):356-369, February, 2014
Paper
D. Grissom and P. Brisk
Fast Online Synthesis of Generally Programmable Digital Microfluidic Biochips
10th IEEE/ACM/IFIP International Conference on Hardware/Software Codesign and System Synthesis (CODES-ISSS)
Tampere, Finland, October 7-12, 2012, pp. 413-422
Paper Slides
Contact
Please direct any questions, comments, or other inquiries to the following e-mail address: microfluidics@cs.ucr.edu
Acknowledgment
This material is based upon work supported by the National Science Foundation under Grant Numbers 1035603, 1536026, and 1545097. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.