Sample Preparation Framework: Overview
Introduction
A fundamental requirement of many microfluidic chips is the need to control the concentration of a fluid. For example, consider a microfluidic chip that uses a sample of blood to diagnose malaria. Blood is a very concentrated fluid, so it is usually necessary to dilute a blood sample before it can be analyzed onchip. However, if the blood sample is diluted too much, the concentration of the malaria antigens present in the blood could drop below the chip’s limit of detection and become undetectable (a “false negative”). So there is an optimal amount of dilution that is required for a microfluidic chip to function, and this optimal amount may vary from sample to sample. This is just one example of the many applications that require precise control of fluid concentration in microfluidics.
Most microfluidic chips rely on human operators to prepare samples and other chemicals at the correct concentrations. A few microfluidic chips are capable of diluting fluids on-chip; for example, a single mixing operation on a Digital Microfluidic Biochip (DMFB) can dilute a fluid to 50% of its starting concentration. By repeating this dilution process multiple times, dilutions of 25%, 12.5%, 6.25%, etc. are also possible. While DMFBs are programmable and can perform virtually any desired dilution, the difficulty of programming them has limited their widespread use. As electrode counts increase, programming their operation by hand grows increasingly difficult. A number of researchers have developed algorithms that automate the programming of DMFBs, thereby eliminating the need to program them by hand. Many of these algorithms have already been incorporated into the UCR Digital Microfluidic Biochip Static Simulator and Compiler.
Sample Preparation Framework
Sample preparation is one of the most common applications (or subroutines of larger applications) used in microfluidics. A number of researchers in recent years have introduced algorithms that automatically generate microfluidic sample preparation programs from a concise description of a desired target product. In terms of the Static Simulator, the output of these algorithms are DAGs called Dilution Graphs that perform the necessary steps to prepare the desired sample(s). In principle, Dilution Graphs are benchmarks that can be used to compare the performance of the different algorithms that have been included in the simulator.
The UCR Digital Microfluidic Biochip (DMFB) Sample Preparation Framework is a collection of algorithms that generate Sample Preparation DAGs in a format that is compatible with the UCR DMFB Static Simulator. The Sample Preparation Framework employs a 1:1 mixing model, which is appropriate for DMFB technology. Under the 1:1 mixing model, each operation mixes two droplets of equal volumes V, producing a single droplet whose volume is 2V; that droplet can immediately be split into two equal sized droplets of volume V. At present, the sample preparation framework and simulator assume perfect spliting; future work will introduce uncertainty into the splitting model.
We plan to periodically update the Sample Preparation Framework with additional algorithms that we introduce ourselves and/or read in the literature. The repository of algorithms is neither complete nor exhaustive, nor do we ever expect it to be. By releasing the source code of both the DMFB Static Simulator and the Sample Preparation Framework, we encourage researchers working on these topics to compare with algorithms introduced by others, and to evaluate the performance of their algorithms in the simulator.
Examples
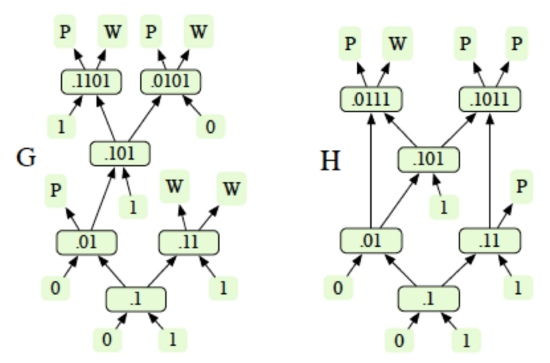
Under the 1-1 mixing model, the concentration of a sample can be represented in binary, e.g., .1 = 50%, .01 = 25%, etc. Samples that are input to the system have an initial concentration of 1 (100%), while the buffer fluid used for dilution has a concentration of 0. Two different dilution graphs are shown below. Each intermediate node is labeled with the concentration of the droplets it generates. Output droplets are labeled as product (P) or waste (W). Only the product droplets are desired.
Objectives
A wide variety of sample preparation algorithms have been introduced, which vary in terms of their objectives. For example, some produce a single droplet of a desired target concentration, while others produce multiple droplets. Additionally, objectives may include:
♦ Minimize the number of mix operations in the Dilution Graph
♦ Minimize reactant usage (i.e., minimize the number of input nodes in the Dilution Graph)
♦ Minimize the number of waste droplets produced
Reference
The UCR Digital Microfluidic Biochip (DMFB) Sample Preparation Framework has been summarized in the following paper. (Similar to this website, the paper simply lists the sample preparation algorithms that have been implemented and provides references to the papers that introduced them). We respectfully request that you cite this papers if you use our binaries or source code in your own work.
D. Grissom, C. Curtis, S. Windh, C. Phung, N. Kumar, Z. Zimmerman, K. O'Neal, J. McDaniel, N. Liao, and P. Brisk
An Open-source Compiler and PCB Synthesis Tool for Digital Microfluidic Biochips
Integration: The VLSI Journal
51:169-193, September, 2015
Paper
Contact
Please direct any questions, comments, or other inquiries to the following e-mail address: microfluidics@cs.ucr.edu
Acknowledgment
This material is based upon work supported by the National Science Foundation under Grant Numbers 1035603, 1536026, and 1545097. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.