Benchmarks
Benchmarks
The three benchmarks listed on this page are widely used to evaluation DMFB compilation and synthesis algorithms. To the best of our knowledge, this information was originally contained in a non-peer reviewed document written by Fei Su and Krishnendu Chakrabarty from Duke University, and hosted on Fei Su's webpage at Duke. The document has since disappeared, so we are re-disseminating the benchmarks here to better serve the research community.
These benchmarks are included with our source code download, along with several others.
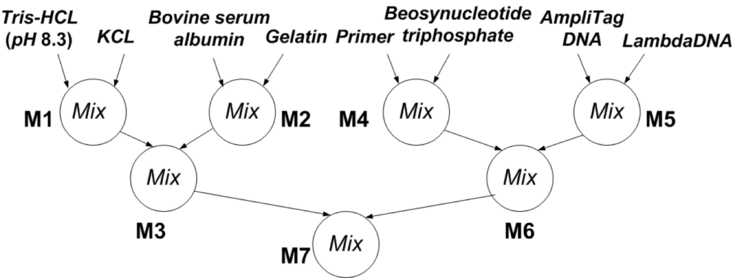
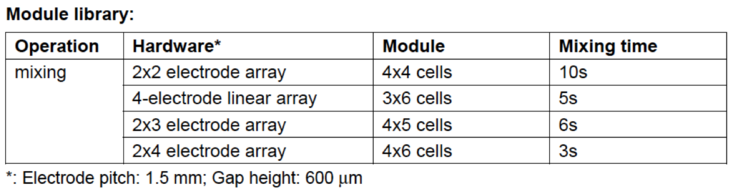
PCR Mixing Tree
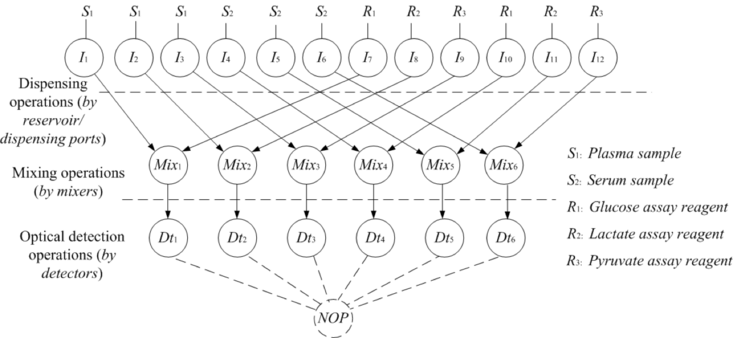
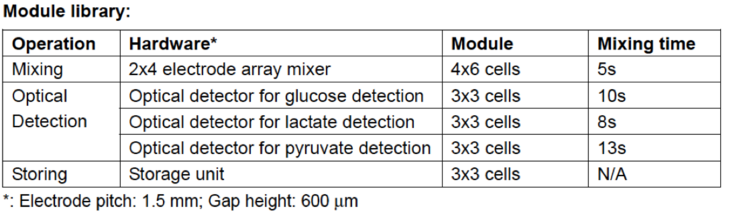
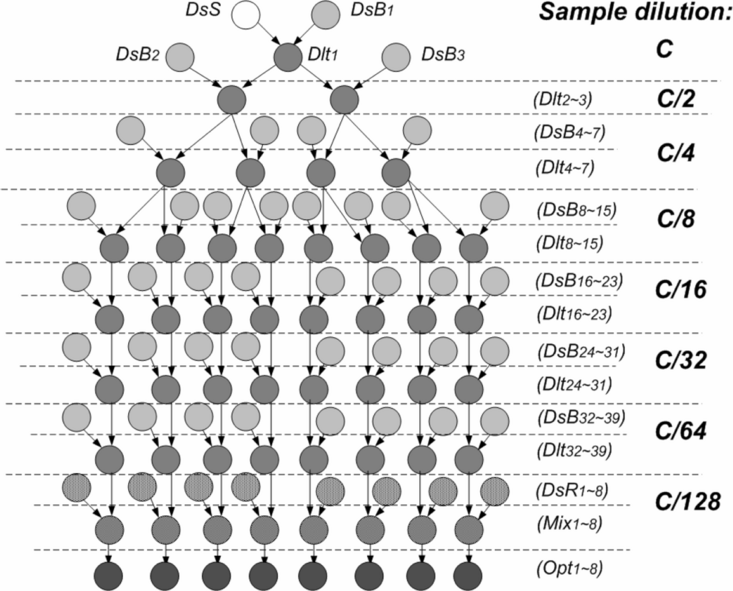
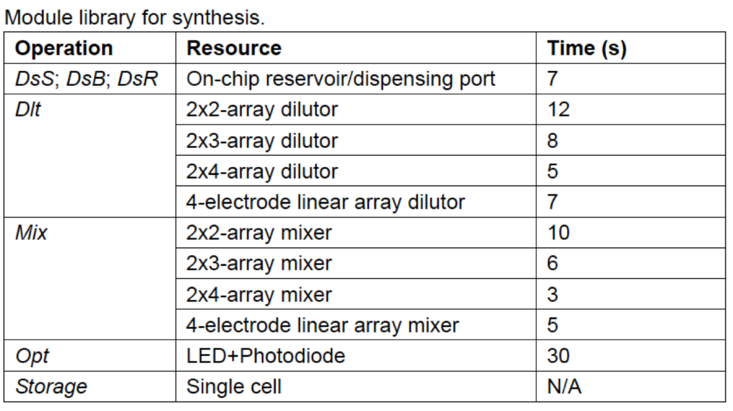
Multiplexed In Vitro Diagnostics Assay
Protein Assay
Contact
Please direct any questions, comments, or other inquiries to the following e-mail address: microfluidics@cs.ucr.edu
Acknowledgment
This material is based upon work supported by the National Science Foundation under Grant Numbers 1035603, 1536026, and 1545097. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.