Static Simulator: Overview
Introduction
The UCR Digital Microfluidic Biochip (DMFB) Static Simulator is a repository of algorithms that compile specifications of biochemical reactions (assays) onto DMFBs. In addition to the compiler, the software supports algorithmic aspects of application-specific specialization and Printed Circuit Board (PCB) design and layout. The framework is designed for modularity and ease-of-use so that researchers can easily implement their own algorithms and compare against the current state-of-the-art. The framework output is graphical, with the primary objectives being: (1) to simplify debugging; and (2) to support visual presentation for academic papers, presentations, etc.
The Static Simulator is built in C++ for speed. All the back-end file I/O, interface standards, etc. are already defined such that a developer only needs to modify a few select functions when designing a new algorithm (e.g. scheduler, placer, router). Timing mechanisms are even built in for fair comparisons. For end-users simply looking to use the simulator for educational purposes or non-developmental purposes, we provide a simple Java GUI interface that calls the C++ simulator. The simulator can also be called directly from the command line if desired.
The key limitation of the Static Simulator is that it does not support real-time feedback from integrated sensors or video monitors. We will shortly release a separate dynamic framework which supports these features.
Static Simulator: Overview
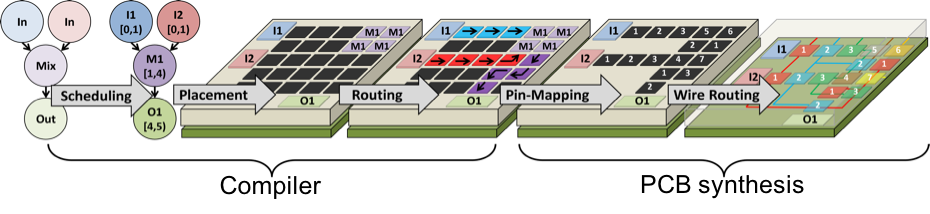
The Static Simulator, at present, consists of five stages, as shown below.
Compiler
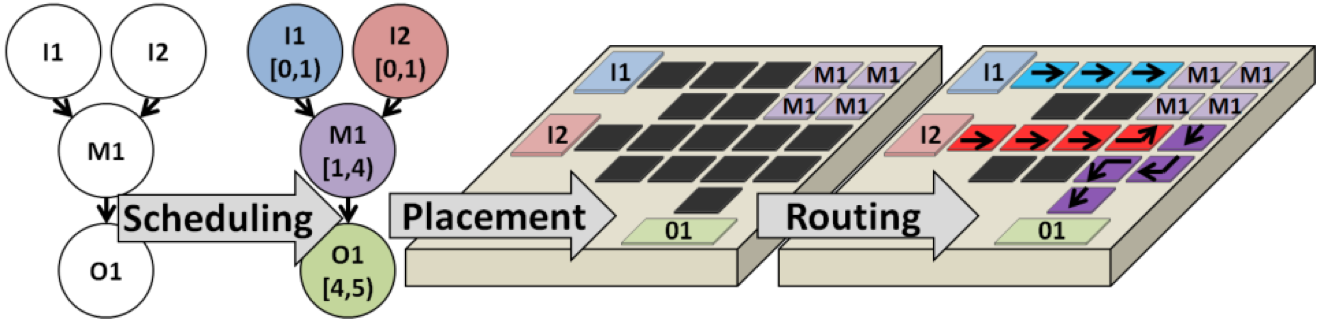
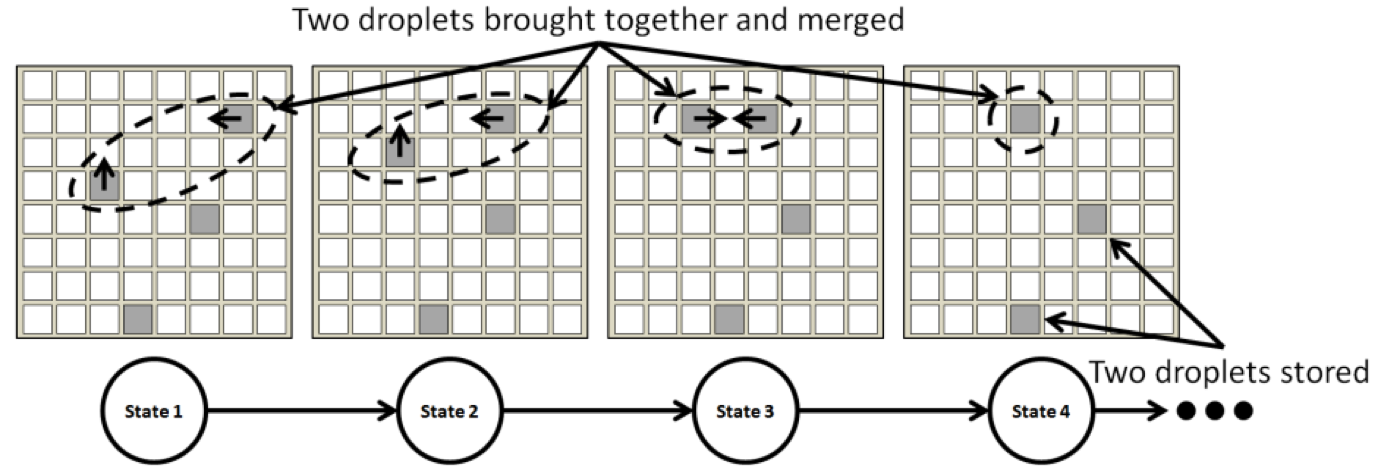
The compiler converts a biochemical assay, specified as a Directed Acyclic Graph (DAG), into an electrode actuation sequence that executes the assay on the DMFB. The key steps to be solved are Scheduling, Placement, and Routing, all of which are NP-complete; the development of algorithms and heuristics to solve these problems has been an active area of research since approximately 2001. The electrode actuation sequence can be viewed as a linear state machine (a Moore Machine) in which each state outputs a subset of electrodes to actuate. The default actuation frequency in the framework is 100 Hz.
PCB Synthesis
The objective of PCB synthesis is to reduce the cost of the DMFB and the PCB onto which it is mounted. At present, we do not automatically generate a workable or usable PCB, although we may extend the framework to do so in the future. The objective here is to collect and disseminate some of the algorithmic underpinnings of such a tool. PCB cost reduction typically entails two interdependent strategies:
♦ Reduce the number of control pins (Pin Mapping); and
♦ Reduce the number of PCB layers (Wire Routing).
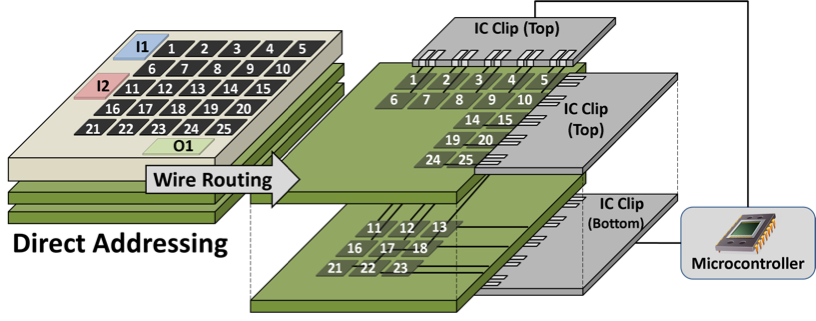
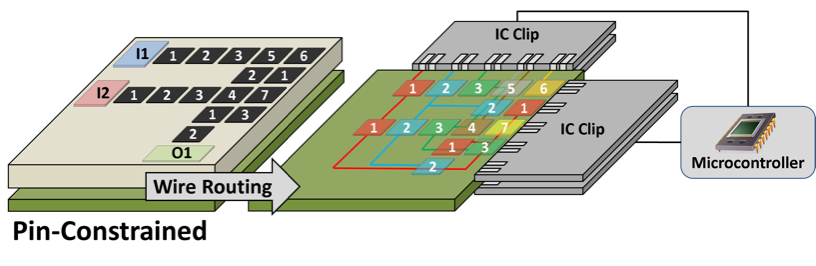
A Direct Addressing DMFB is an MxN array of independent electrodes, each of which is driven by a dedicated control pin on the perimeter of the chip.
By examining the electrode actuation sequence, several opportunities may arise to reduce the number of control pins by making the device application-specific:
♦ Remove electrodes (and their associated control pins) that are never actuated
♦ A Pin-constrained DMFB allows electrodes with compatible actuation sequences to share a common control pin.
The wire routing stage instantiates connections between each control pin on the perimeter of the chip and the group of electrodes that it drives. This involves solving two interdependent problems: electrode group partitioning and escape routing. The partitioner determines which electrode groups are routed on each PCB layer; for each layer, the escape router creates wire routes from each control pin to the group of electrodes that it drives on each layer. Many papers have also been published that co-optimize pin-mapping and wire routing, and we have re-implemented several of the algorithms within our framework.
Reference
The UCR Digital Microfluidic Biochip (DMFB) Static Simulator has been described in detail in the following two papers. We respectfully request that you cite both of these papers if you use our binaries or source code in your own work.
D. Grissom, C. Curtis, S. Windh, C. Phung, N. Kumar, Z. Zimmerman, K. O'Neal, J. McDaniel, N. Liao, and P. Brisk
An Open-source Compiler and PCB Synthesis Tool for Digital Microfluidic Biochips
Integration: The VLSI Journal
51:169-193, September, 2015
Paper
D. Grissom, K. O'Neal, B. Preciado, H. Patel, R. Doherty, N. Liao, and P. Brisk
A Digital Microfluidic Biochip Synthesis Framework
20th IFIP/IEEE International Conference on Very Large Scale Integration (VLSI-SoC)
Santa Cruz, CA, USA, October 7-12, 2012 pp. 177-182
Paper Slides
The VLSI-SoC 2012 paper described a preliminary version of the framework with fewer algorithms and more limited capabilities. If you are space constrained and can only fit one reference, we prefer that you reference the Integration paper.
Contact
Please direct any questions, comments, or other inquiries to the following e-mail address: microfluidics@cs.ucr.edu
Acknowledgment
This material is based upon work supported by the National Science Foundation under Grant Numbers 1035603, 1536026, and 1545097. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.